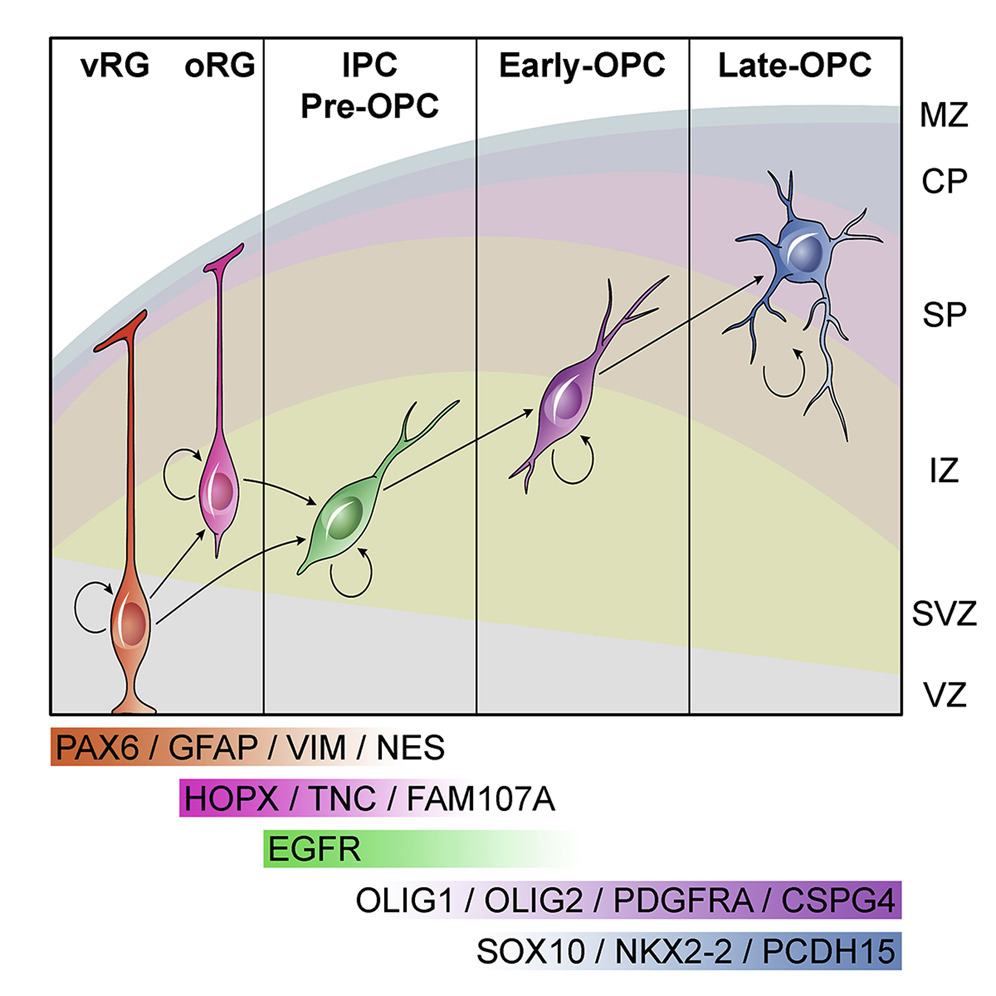
Using single-cell RNA sequencing of immunopanned oligodendrocyte precursor cells (OPC), we identified a novel pre-OPC population that is generated from outer radial glia cells and is marked by EGFR. We show that this population requires EGFR mediated signaling to produce oligodendrocytes. Because this pre-OPC cell type comes from outer radial glia cells which are enriched in humans, these cells may play a role in cortical expansion.
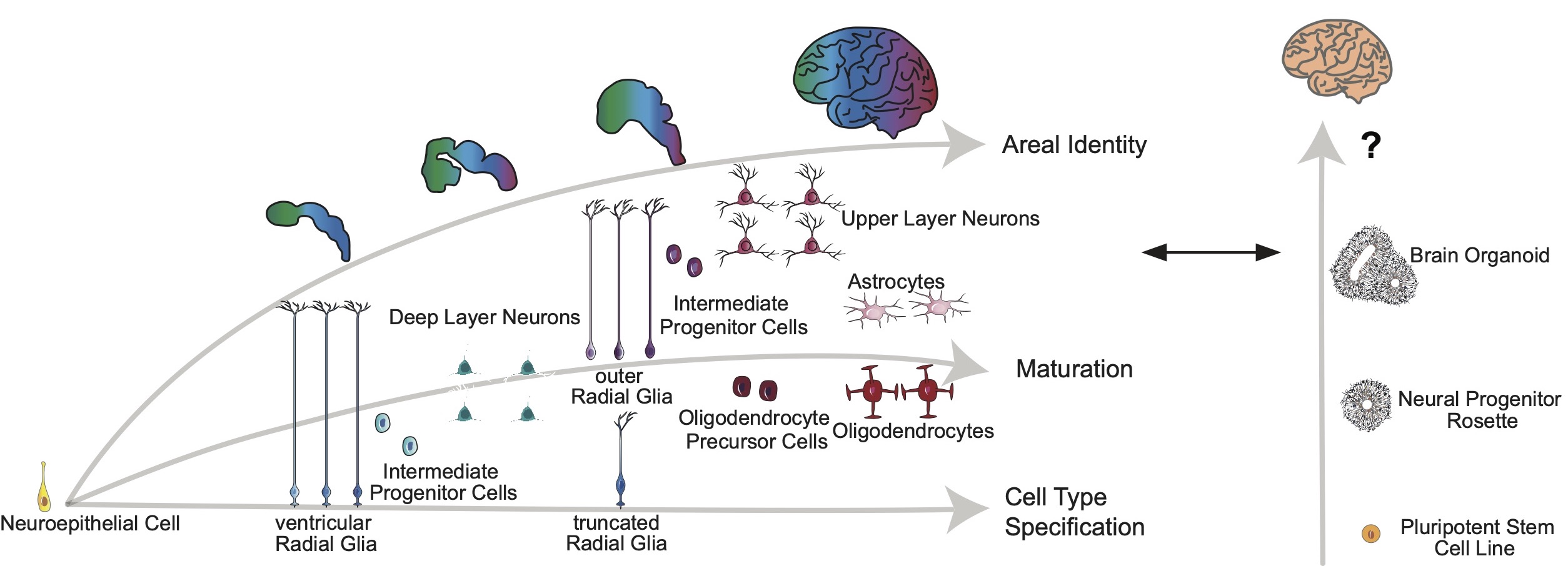
We compared cell types in cortical organoid systems and the primary developing human brain using single-cell RNA sequencing. We identify that major cell types are preserved across both systems, but that cortical organoids suffer from impaired cell subtype specification, incongruent maturation, and areal heterogeneity. We observe that organoids have higher levels of metabolic stress and that activation of this stress in primary cells in culture also impairs subtype specification, while transplantation of organoids into the mouse brain alleviates stress and improves subtypes. These data suggest that organoid cultures can be improved to be more physiologically relevant, but also demonstrates that the limitations of cell type specification are able to be resolved.
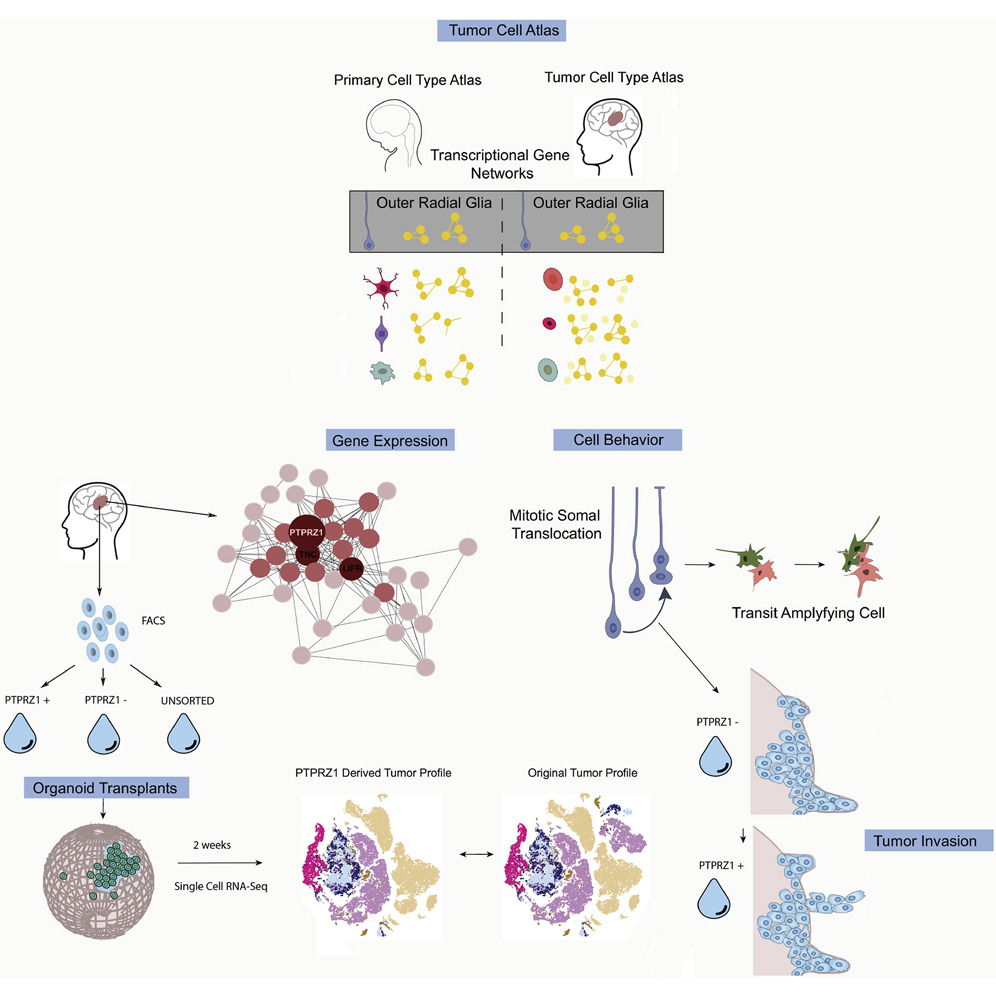
To understand the developmental origins of glioblastoma, we collected single-cell RNA sequencing from 11 primary tumors and compared it to our developmental datasets. We identified the reactivation of the outer radial glia cell, and observed it was undergoing its classic "jump and divide" mitotic somal translocation behavior that may be mediated tumor invasion via PTPRZ1. Using a novel tumor transplantation assay into cortical organoids, we saw these cells were capable of giving rise to neuronal and glial tumor populations.
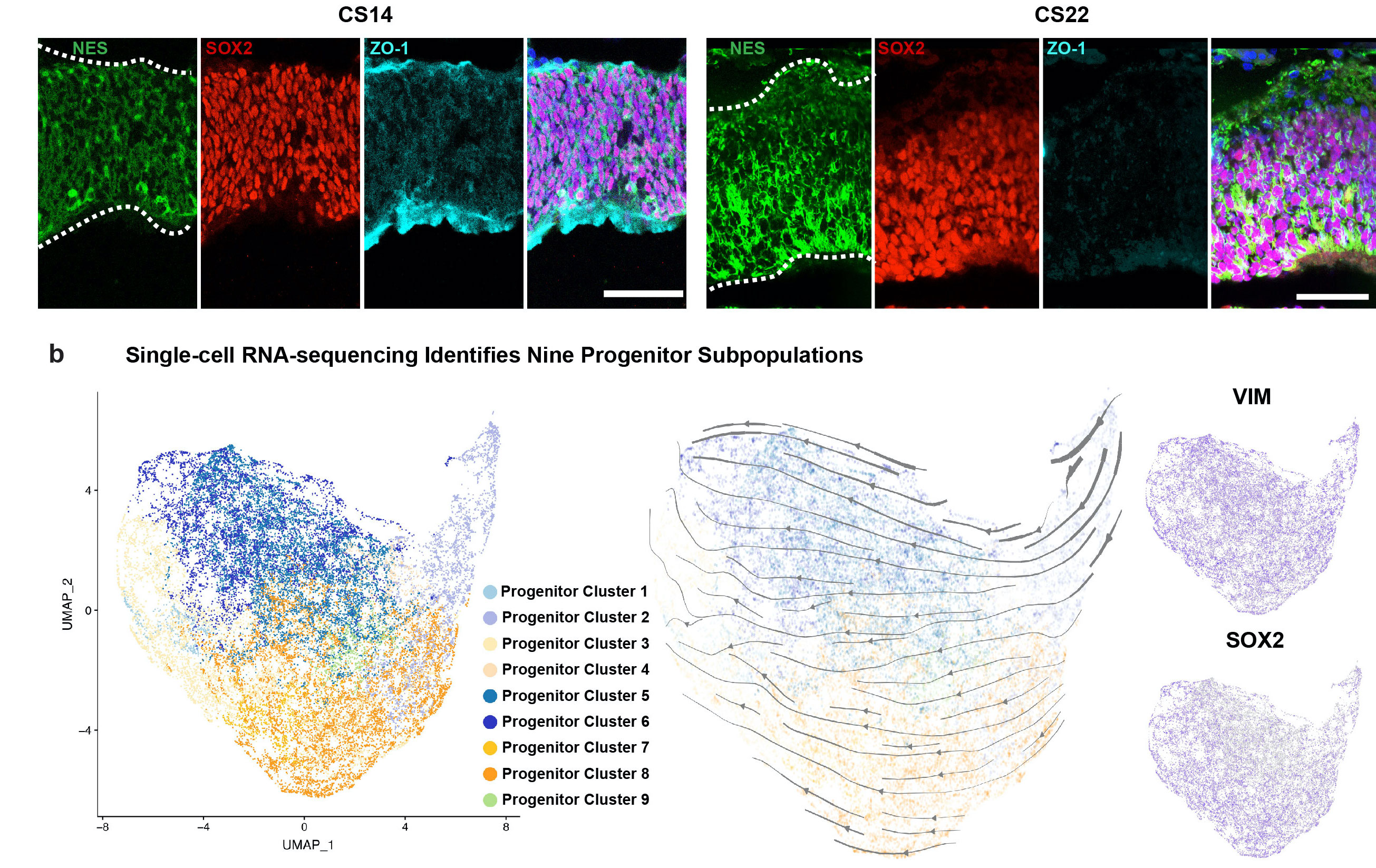
We generated an atlas of difficult to access early timepoints of human brain and cortical development. In the developing forebrain, we identify nine progenitor populations that span neuroepithelial, radial glia, and mesenchymal progenitors including previously undescribed populations. We pair this single-cell analysis with an immunostaining atlas of these populations across the whole brain. We highlight two populations that are enriched in the human at these timepoints but are not represented at parallel developmental stages in the mouse.
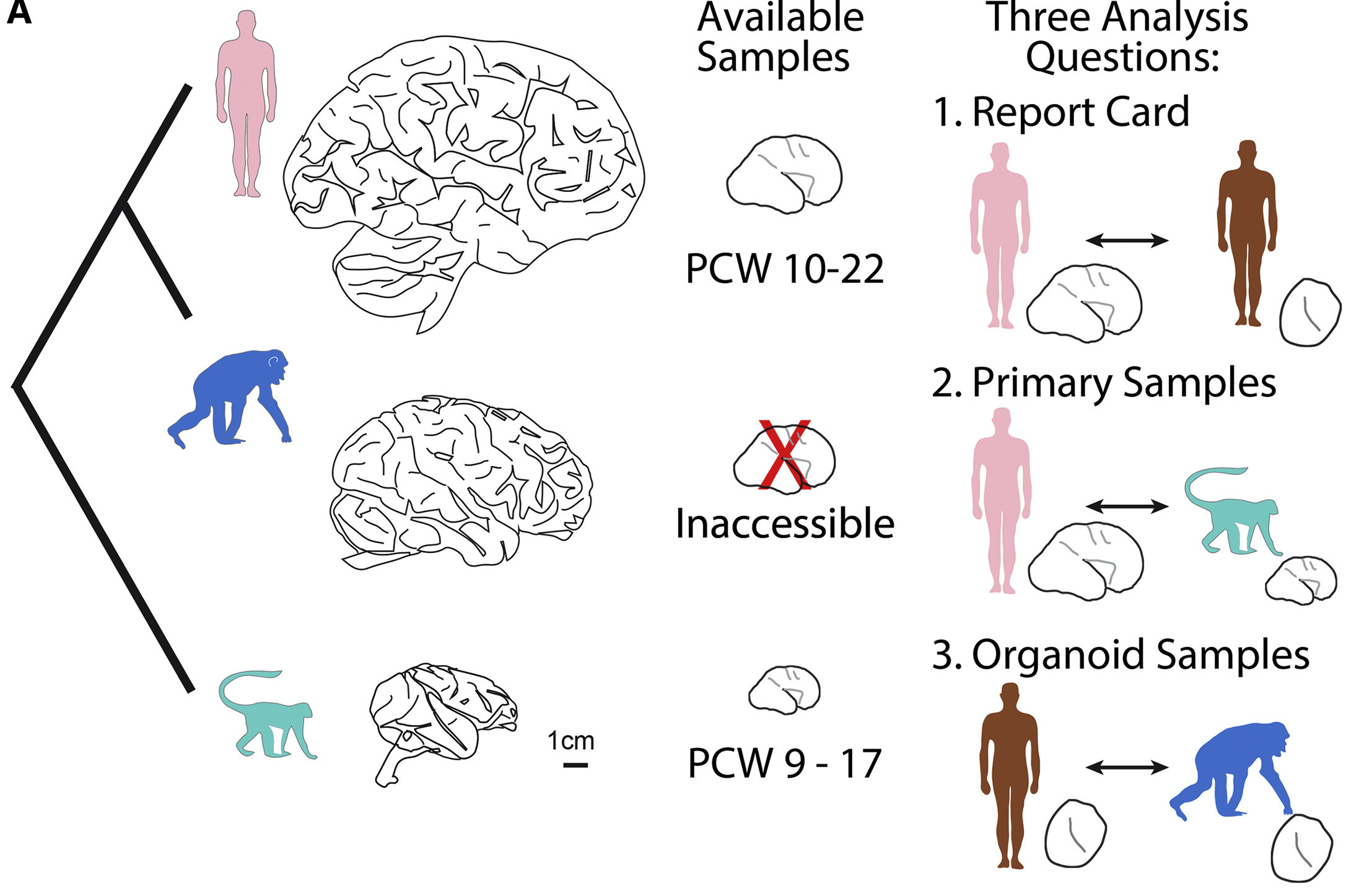
The human cortex is substantially expanded compared to our closest relatives, and these differences emerge at birth. Using primary human and macaque developing cortex, as well as cortical organoids from human and chimpanzee cells, we probed these differences using single-cell RNA sequencing. We identified major cell populations conserved across all systems, and highlight several hundred genes that are differentially expressed between human and non-human primates. These genes highlighted mTOR signaling to be up-regulated in the human cortex, and we find this activation is specific to the outer radial glia cells, suggesting cell type specific pathway activation may contribute to cortical expansion.
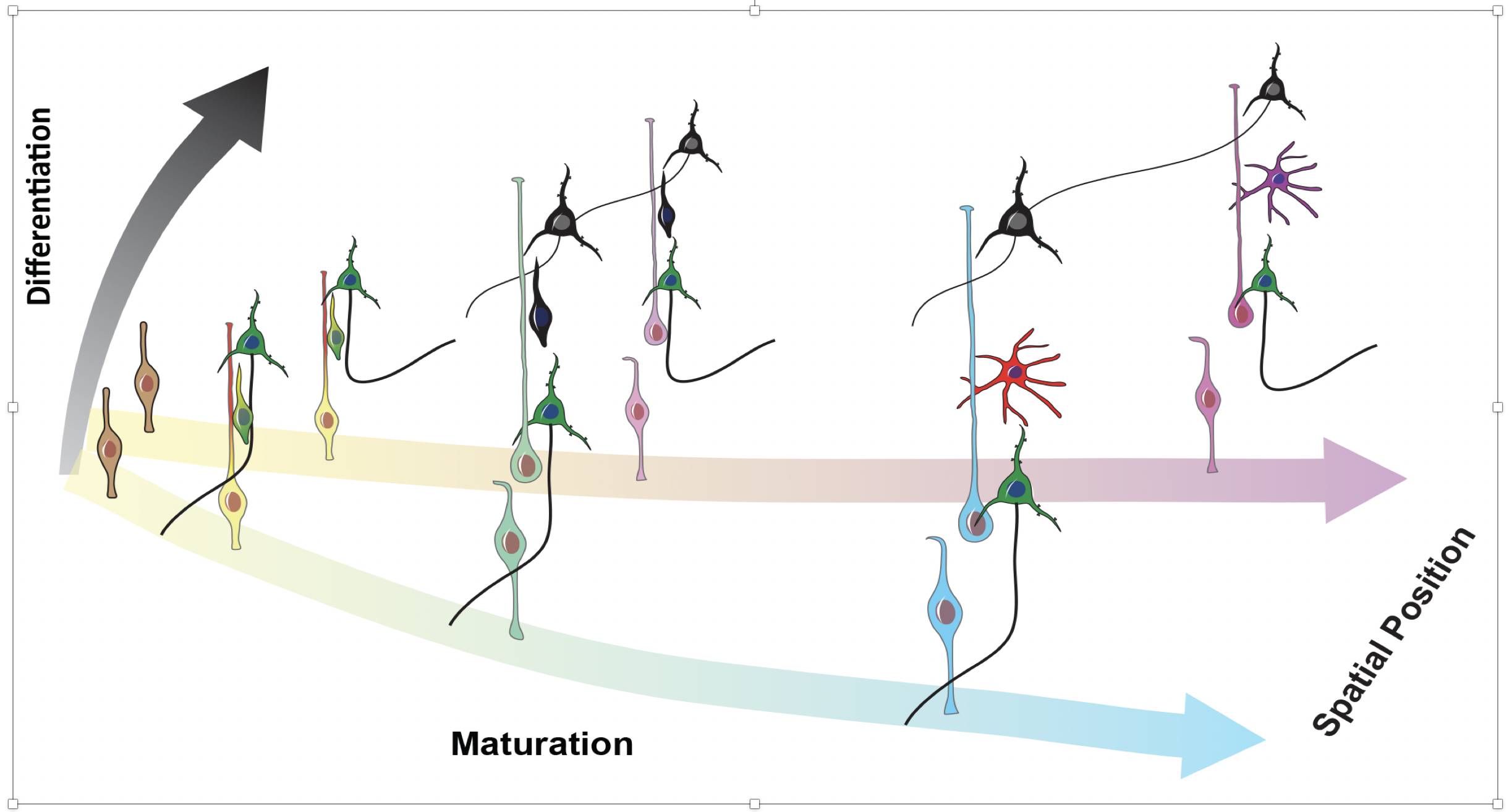
The human cortex is substantially expanded compared to our closest relatives, and these differences emerge at birth. Using primary human and macaque developing cortex, as well as cortical organoids from human and chimpanzee cells, we probed these differences using single-cell RNA sequencing. We identified major cell populations conserved across all systems, and highlight several hundred genes that are differentially expressed between human and non-human primates. These genes highlighted mTOR signaling to be up-regulated in the human cortex, and we find this activation is specific to the outer radial glia cells, suggesting cell type specific pathway activation may contribute to cortical expansion.