Overview
During a normal day, our body undergoes many changes in metabolism, which requires rapid adaptation to different conditions. For example, after a meal, our cells transition from seeing minimal levels of nutrients such as glucose or lipids, to suddenly having a large influx of these metabolites. These cells have to rapidly adapt to these large environmental changes to maximize their ability to take up, utilize, or store these metabolites for survival. Failure to adapt to these changes is linked to many metabolic diseases, such as obesity and fatty liver. Our lab is interested in discovering novel molecular mechanisms that regulate adaptive metabolic processes using a combination of big-data, physiology, genetics, and biochemistry. So...we like to hunt for genes, and find a new function to them!
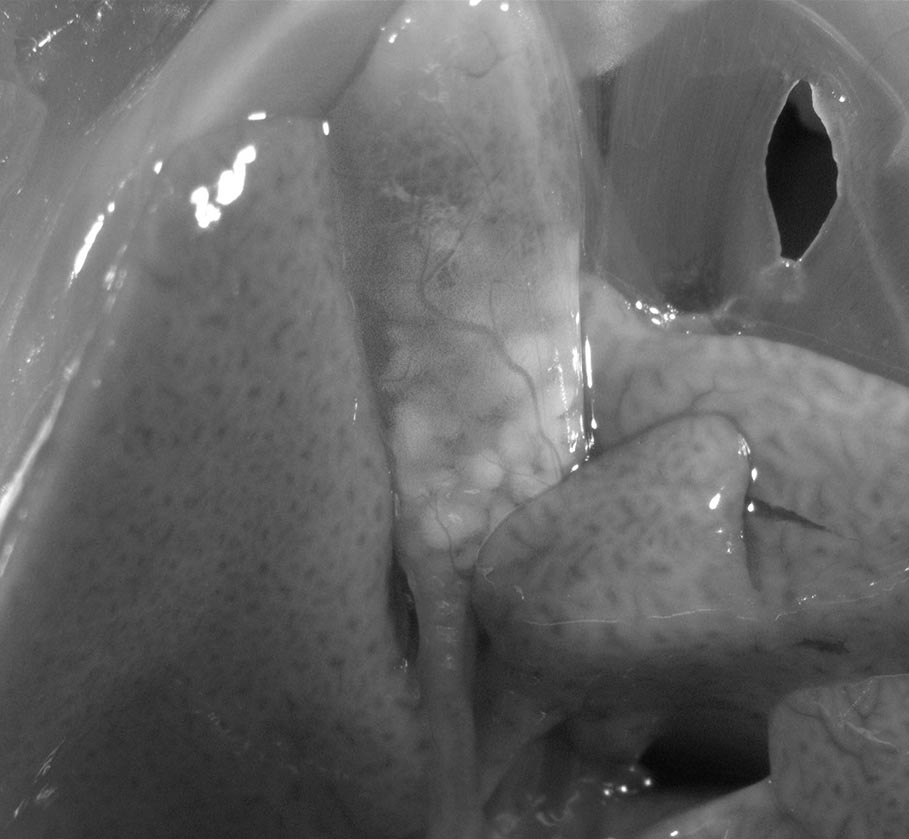
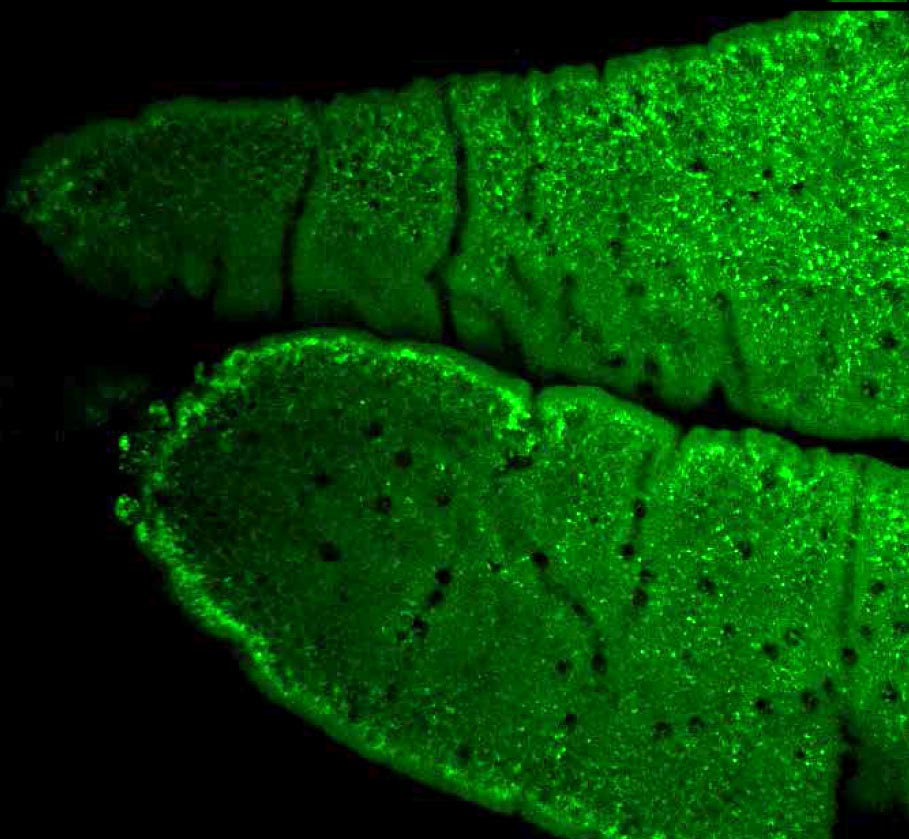
Lipid Accumulation in the Intestine
Role of FXR in Regulating Lipid Metabolism
Rapid adaptation requires turning many genes on, a process that is regulated by transcription factors. In our lab, we study a specific transcription factor that is a member of the nuclear receptor superfamily, called Farnesoid X Receptor (FXR). FXR is a ligand-activated transcription factor, meaning that it regulates genes when a ligand/nutrient, in this case a bile acid, binds to the receptor. Bile acids are multi-faceted lipids that can act both as signaling molecules and detergents that facilitate lipid absorption in the intestine. FXR regulates gene programs that maintain bile acid homeostasis, but also control lipid metabolism, particularly triglyceride metabolism in the liver and intestine, as well as regulating hepato-protective and anti-inflammatory pathways. We are interested in understanding the many FXR-regulated pathways in search of novel mechanisms that control metabolism.
Metabolic Control via mRNA Decay
Just as metabolic adaptation requires gene programs to be activated and turned on, there are pathways that control turning genes off. We identified an RNA-binding protein (RBP) family (Zfp36 or Tis11) as gate-keepers of these so-called "off-switches". In the presence of a stimulus, these RBPs are rapidly activated, and they in turn target mRNAs in a sequence-dependent manner for degradation. We have identified different conditions that regulate the expression of these RBPs, including FXR, and we are now working towards identifying pathways that are regulated at the level of mRNA stability. One pathway we have already identified is bile acid synthesis. We have shown that FXR induces ZFP36L1 which then signals to degrade the rate-limiting enzyme of bile acid synthesis, CYP7A1. We are currently investigating how ZFP36 family members degrade targets using biochemical approaches, and what pathways they target using mouse physiology experiments coupled to big data analysis.
Using Systems Biology to Identify Novel Pathways that Control Lipid Metabolism
Our genomes contain over 25,000 genes, yet we probably only understand the function of 20-25% of these genes. How can we identify a function for the remaining uncharacterized genes? We have exploited the natural variaton in a panel of mice and integrated proteomic and lipidomic analyses in the livers of over 100 strains to make a rich resource that can be profiled to find new functions for individual genes. We are also expanding our resource with additional analyses, including machine learning, to uncover more complex biology and interactions.