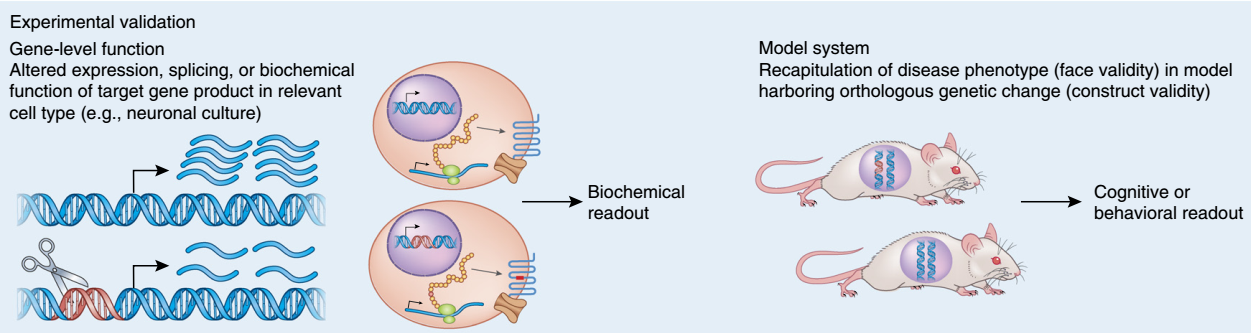
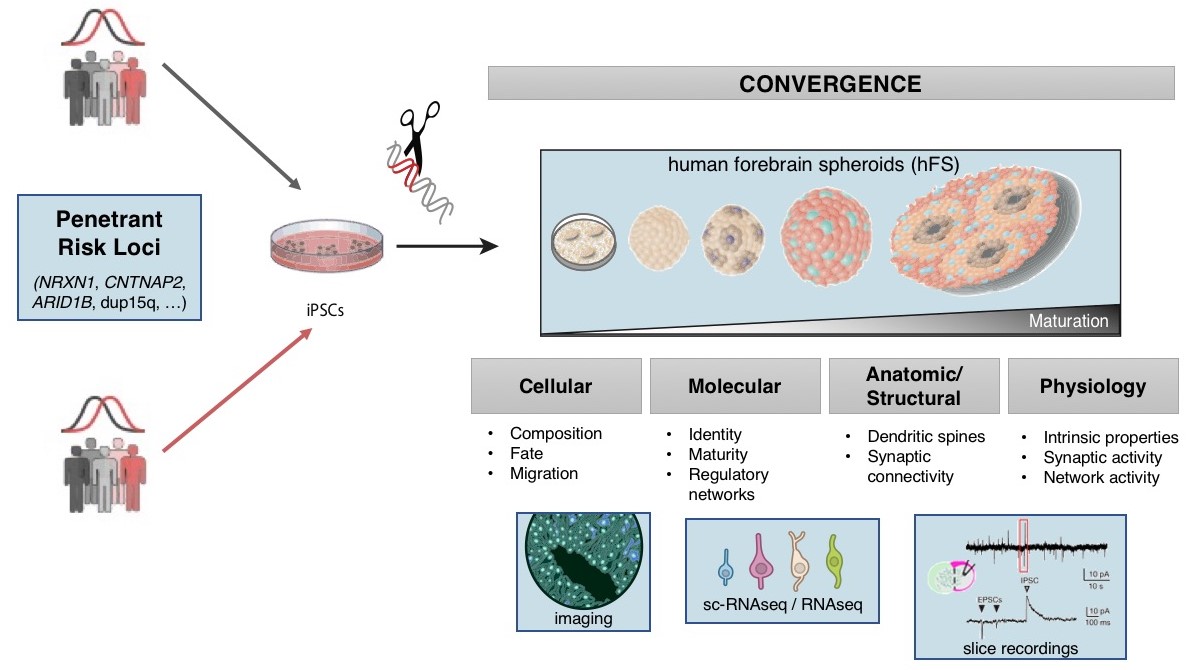
Once the genes implicated in a disorder are identified, understanding the mechanism(s) by which they lead to disease is approachable. We use genetic engineering in model systems such as rodents (in vivo), and human neural stem cells (in vitro) to understand the biological impact of risk variants (schematic shown below).
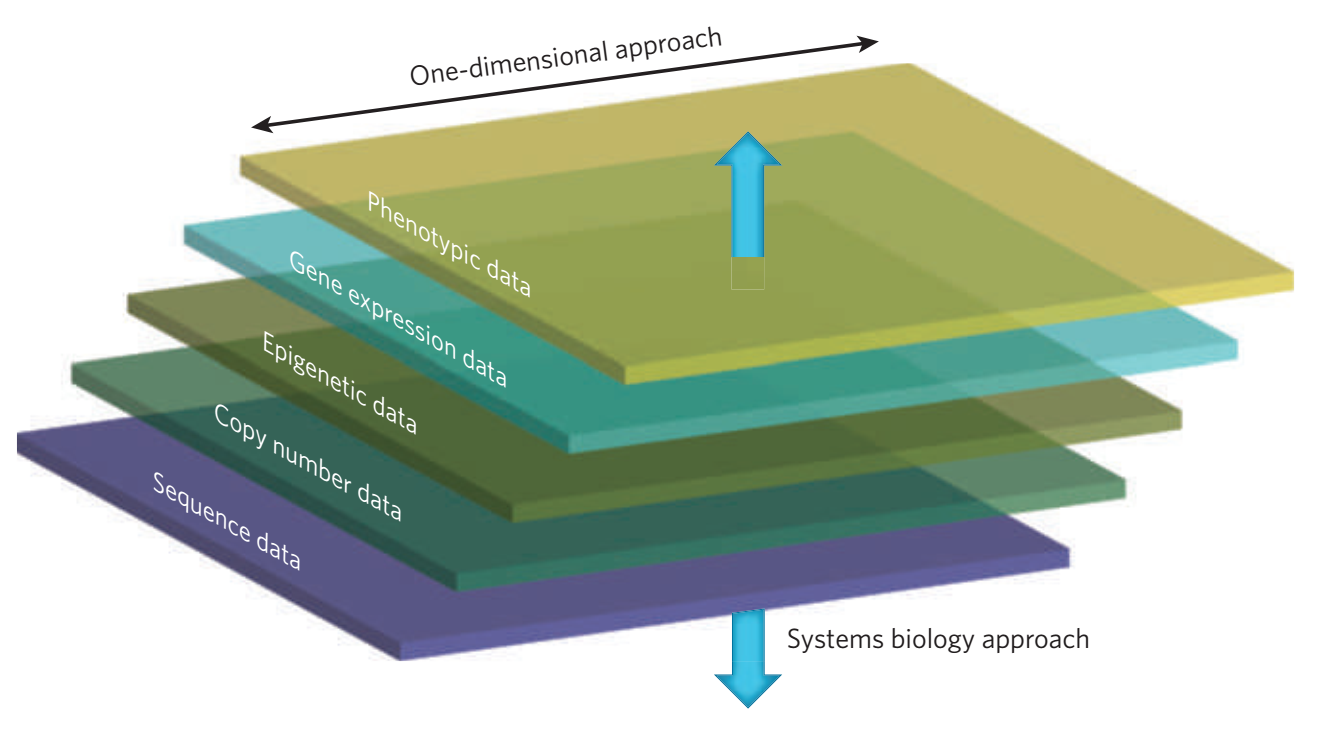
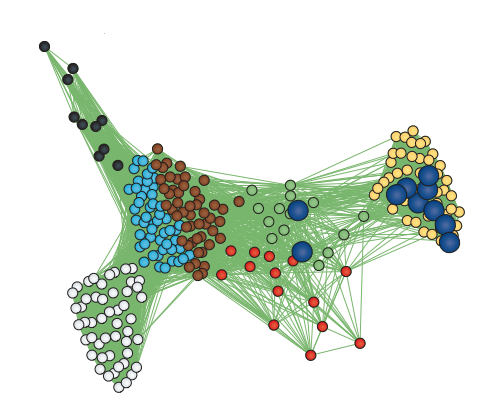
However, since genes do not act in isolation and many complex human disorders involve the interaction of many genes, we also rely on systems biology methods to understand the biological pathways that may be impacted by disease associated variation. We employ multi-dimensional approaches to link genotype to phenotype by profiling genome-wide measures such as whole genome sequencing, transcriptomics, and epigenetics. This has necessitated the development and application of gene network methods, which form the backbone of our systems biology framework.
See also:
Gandal
et al. 2016,
The road to precision psychiatry: translating genetics into disease mechanisms,
Nat Neurosci.
Geschwind and Konopka 2009,
Neuroscience in the era of functional genomics and systems biology,
Nature.
Parikshak
et al. 2015,
Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders,
Nat Rev Genet.
de la Torre-Ubieta
et al. 2016,
Advancing the understanding of autism disease mechanisms through genetics.,
Nat Med.
Pa?ca
et al. 2015,
Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture,
Nat Methods.
Peñagarikano
et al. 2011,
Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits,
Cell. (watch the video abstract here)
Peñagarikano
et al. 2015,
Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism,
Sci Transl Med.
Chandran
et al. 2017,
Inducible and reversible phenotypes in a novel mouse model of Friedreich's Ataxia,
eLife.