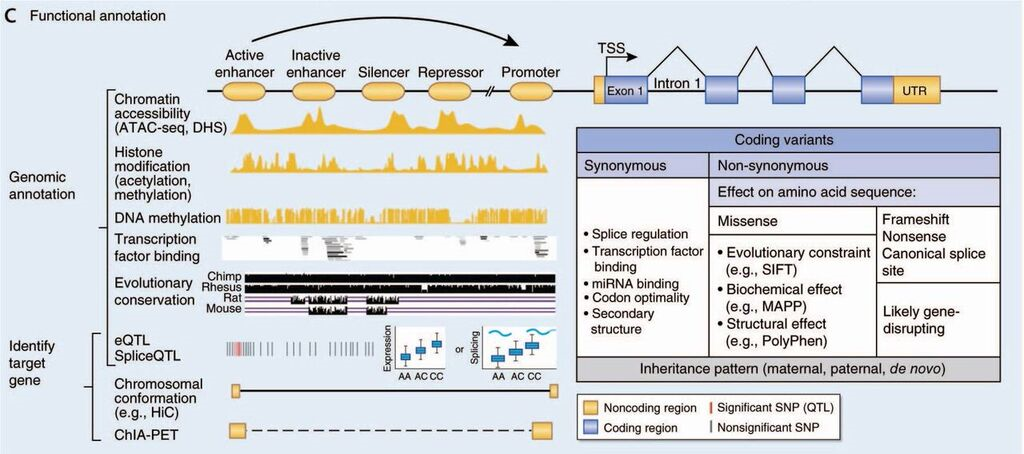
Because a substantial proportion of gene regulation is tissue or stage specific, a crucial component of our work is to build brain relevant maps of these regulatory relationships (illustrated below) by integrating data from many sources including genetic variants, transcriptomics, chromatin accessibility, epigenetic marks, transcription factor binding and 3-dimensional chromatin folding across human development and understand how these regulatory interactions may or may not be conserved in model organism as a leader in the
PsychENCODE project, a large scale NIMH collaborative effort to map out regulatory elements implicated in psychiatric disease. Since most human genetic variation associated with disease lies in noncoding regions, understanding these interactions is a critical step in modern genomics.
We are starting to build these regulatory maps at the single cell level so as to obtain a detailed circuit level understanding to refine our understanding of normal functioning and disease mechanisms.
See also:
Won
et al. 2016,
Chromosome conformation elucidates regulatory relationships in developing human brain,
Nature.
De la Torre-Ubieta
et al. 2018,
The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis,
Cell.
The PsychENCODE Consortium
et al. 2015,
The PsychENCODE project,
Nat Neurosci.
Gandal
et al. 2016,
The road to precision psychiatry: translating genetics into disease mechanisms,
Nat Neurosci.